A dominant haplotype of the delta variant of A SARS-CoV-2 virus that had been rapidly transmitting in Assam by evading the immune system of humans has revealed the dynamics of viral haplotype transmission in a regional population and novel secrets of viral evolution. The study which shows that this particular variant, though detected in other areas, spread rapidly particularly among the Assamese population, can help developing models for future prediction of the disease at the local level.
The Covid-19 pandemic had caused over 3 million deaths worldwide. The virus mutated into several Variants of Concern (VOCs) that showed a pattern of invasion in specific populations or countries and several variants have been reported from different parts of the world.
A study conducted by a team of scientists comprising of Prof. Ashis Mukherjee, Director, Dr. Mojibur Rohman Khan, Associate Professor, Dr. Maloyjo Joyraj Bhattacharjee, Project Scientist and Mr. Anupam Bhattacharya, Senior Research Associate from the Institute of Advanced Study in Science and Technology (IASST), Guwahati in collaboration with Prof. Wen-Hsiung Li of Academia Sinica Taipei, Taiwan and University of Chicago, USA novel found that haplotype of the delta variant of SARS-CoV-2 underwent selective sweep in Assam in 2021.
The research published in Virology Journal was based on a comprehensive genomic analysis of SARS-CoV-2 samples from nasopharyngeal swabs of COVID-19 positive individuals in Assam. The team sequenced 91 SARS-CoV-2 samples by next-generation sequencing (NGS). Comparative analysis of the genomes from Assam with those from outside revealed that a specific haplotype of the delta variant carrying 13 mutations underwent selective sweep and transmitted very fast in the Assam population. This process was facilitated by a mutation in ORF8, which is involved in immune evasion.
The team further analyzed the expression pattern of the viral genes among the patients and observed a negative correlation of the Ct value of qRT-PCR of the patients with abundant ORF6 transcripts, suggesting that ORF6 can be used as a marker for estimating viral titer. The team concluded that this study revealed an understudied mechanism and dynamics of viral haplotype transmission in a regional population may aid in tracking the viral evolution for better disease management strategy.
Publication Link: DOI: 10.1186/s12985-023-02139-3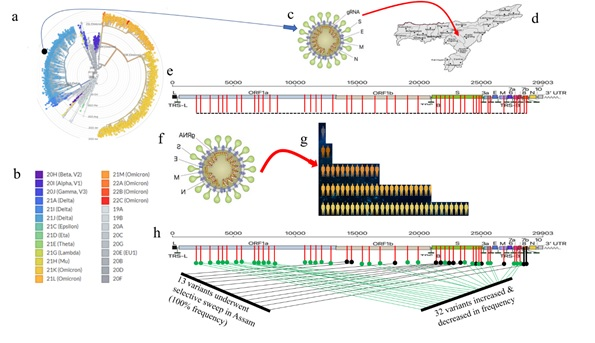
Figure: Model of selective sweep of a SARS-CoV-2 haplotype in Assam population. Globally the SARS-CoV-2 isolates phylogenetically clusters distinctly (a) and are designated with distinct cluster names or variants (b). The study shows a haplotype of delta variant first appeared in Assam (c & d), which altogether carried 45 amino acid variants in the genome (e), and as the haplotype successively spread in Assam population (f & g), 13 variants underwent selective sweep and 32 variants slightly increased and decreased in frequency. This altogether reveals that a haplotype of the delta variant of SARS-CoV-2 carrying 13 amino acid variants underwent selective sweep in the Assam population.